S-BIAD531
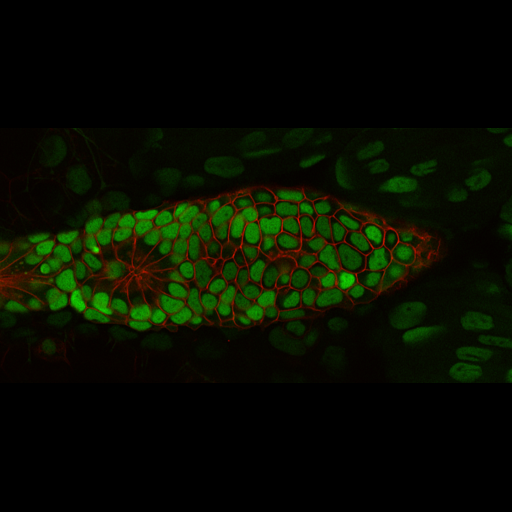
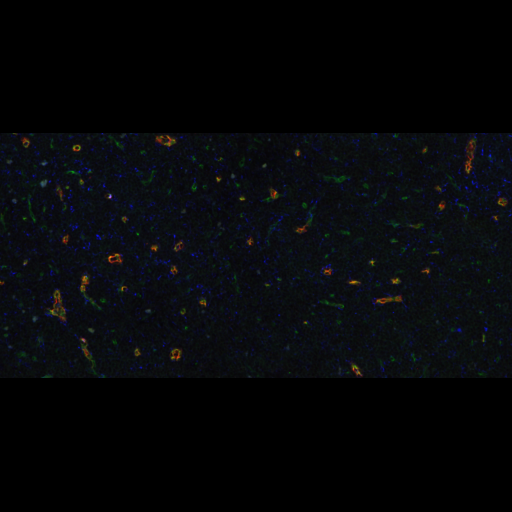
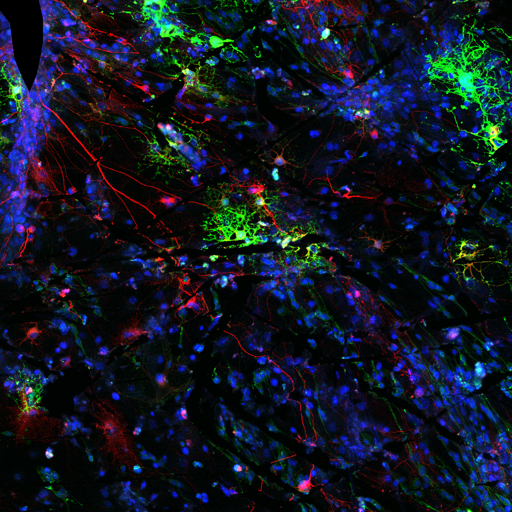
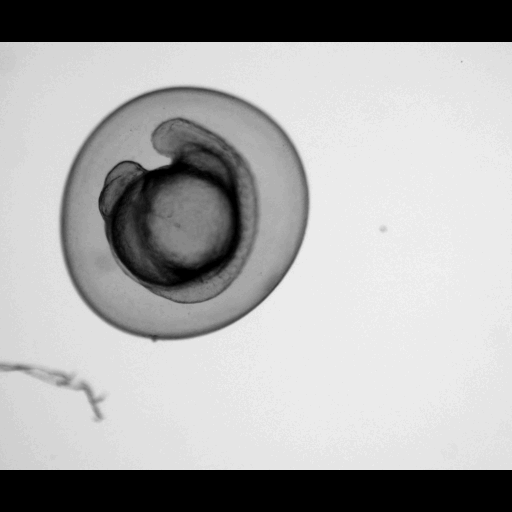
A machine learning-based classifier trained to detect temporal developmental differences across groups of zebrafish embryos, using images from a standard live imaging widefield microscope.
1142 images , 1062 annotations
Tags:
time series, object classification, training data, pixel classification, test data, segmentation, Time series