S-BSST410alpha
Axis Specification in Zebrafish Is Robust to Cell Mixing and Reveals a Regulation of Pattern Formation by Morphogenesis
Released 2020-06-05Timothy Fulton, Benjamin Steventon
Content
- 16 images

Organism
Danio rerio (Zebrafish)
Imaging type
confocal fluorescence microscopy
License
CC0
Viewable images
All images
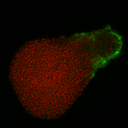
<| Image ID | Filename | Download Size | Actions | IM1 | Figshare Upload/2018-09-28 Krox20 Cdx4_2.tif | Unavailable | Download | IM2 | Figshare Upload/2019-07-10 Pescoids BMP4 647 DKK1b 546 Chordin 488_6hpc_2019_07_10__12_39_25.lsm | Unavailable | Download | IM3 | Figshare Upload/2019-07-26 pH3 647 Eomes 488 Ntl 546 Timecourse_1hpc_p5.lsm | Unavailable | Download | IM4 | Figshare Upload/2019-07-26 pH3 647 Eomes 488 Ntl 546 Timecourse_2hpc_p1.lsm | Unavailable | Download | IM5 | Figshare Upload/2019-07-26 pH3 647 Eomes 488 Ntl 546 Timecourse_4hpc_p2.lsm | Unavailable | Download | IM6 | Figshare Upload/2019-07-26 pH3 647 Eomes 488 Ntl 546 Timecourse_7hpc_p3.lsm | Unavailable | Download | IM7 | Figshare Upload/2020-01-09 Nanos 647 Sox32 546 Vasa 488_7hpc_2020_01_09__21_42_30__p2.lsm | Unavailable | Download | IM8 | Figshare Upload/2020-01-09 Tbxta 647 Sox2 546 Sox17 488_7hpc_2020_01_09__11_29_14__p1.lsm | Unavailable | Download | IM9 | Figshare Upload/2020-01-10 Wnt11 546 Frd 647_7hpc_2020_01_10__15_09_58__p1.lsm | Unavailable | Download | IM10 | Figshare Upload/2020-01-16 BMP2b (488) BMP4 (647) BMP7 (546)Timecourse_2hpc_2020_01_17__08_53_05__p8.lsm | Unavailable | Download | IM11 | Figshare Upload/2020-01-16 BMP2b (488) BMP4 (647) BMP7 (546)Timecourse_5hpc_2020_01_16__16_54_24__p2.lsm | Unavailable | Download | IM12 | Figshare Upload/2020-01-16 BMP2b (488) BMP4 (647) BMP7 (546)Timecourse_7hpc_2020_01_16__14_12_19__p2.lsm | Unavailable | Download | IM13 | Figshare Upload/2020-02-26 TCFGFP Timecourse HCR_2hpc_p12.lsm | Unavailable | Download | IM14 | Figshare Upload/2020-02-26 TCFGFP Timecourse HCR_5hpc_p22.lsm | Unavailable | Download | IM15 | Figshare Upload/Pescoid_8hpc_Keratin647_2.lsm | Unavailable | Download | IM16 | Figshare Upload/TCF647_7hpc.tif | Unavailable | Download |
|---|